Spotlight on human dental pulp: With a snapshot of more than 4,300 proteins for the Human Proteome Project, the Overall Lab increases the number of proteins known to exist in teeth.
A team of UBC Dentistry researchers led by Prof. Chris Overall (www.clip.ubc.ca), including Drs. Ulrich Eckhard, Simon Abbey (UBC Dentistry MSc student), Ian Mathew (oral surgeon), Giada Marino and Grace Tharmarajah, recently published a study focused on human dental pulp.
The dental pulp is the “nerve”, lying within the mineralized outer covering of the tooth. Using contemporary mass spectrometry technology in their lab, these researchers identified more than 4,300 proteins, representing nearly 22 percent of predicted human proteins, including 17 proteins predicted from genes but that have never been observed before1.
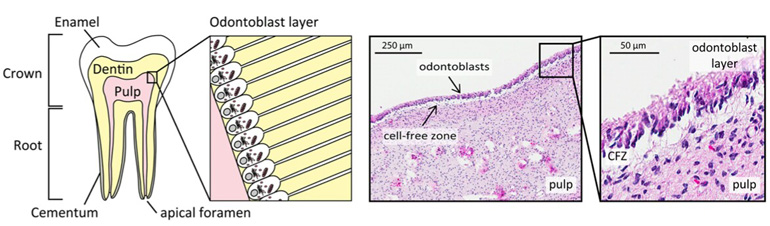
This study, the most comprehensive snapshot of any dental tissue so far, was featured in an editorial of the Human Proteome Project Special Issue published in the prestigious Journal of Proteome Research for the 14th Human Proteome Organization World Congress in 20152. Furthermore, the study revealed that specialized enzymes called proteases that cut proteins are highly important to maintain tissue function: Over 9,000 cut protein ends (termini) were identified in pulp, revealing pervasive protease action in a healthy human tissue.
For many years, studies focused on the role of proteases as non-selective protein degraders. However over the last 15 years, Overall and his laboratory have pioneered the idea that proteases can act as sharp and highly specific scissors that precisely cut, or process, proteins rather than just degrading them. Such finely tuned processing can generate new protein products with altered protein termini and thus different protein functions and localization in cells and tissues3. For example, protein activity can be turned on or off by proteases, which can switch whole signaling cascades on or off, making proteases and their action the focus of many biological processes in health and disease such as gum disease (periodontitis) and in dental pain4.
As a first step, researchers in the Overall Lab used healthy human tissues such as platelets5, erythrocytes6, and now dental pulp1,7 to capture human protein termini to build a solid understanding of proteolytic processing in human health. Thus the lab actively participates in the Chromosome-centric Human Proteome Project (C-HPP; www.c-hpp.org), a global initiative designed by the Human Proteome Organization (www.hupo.org) to map every single protein in the human body (also called human proteome), a quest that began as soon as the Human Genome Project was completed in 20038. Research teams around the world are trying to describe the structure and function of the human proteome and its changes in disease.
In the Overall Lab, the next stage is to analyze diseased samples to identify disruptions in proteolytic processing and signaling that may cast light on new drug targets. Dental pulps from healthy teeth will be compared with pulps from teeth with various degrees of tooth decay to identify key drivers of caries and potential drug candidates to promote tooth dentine repair after caries has been arrested or the tooth restored. In parallel, human gum tissue will be used to study periodontitis, a serious gum infection damaging the soft tissue and destroying the bone supporting the tooth, frequently leading to tooth loss. With this research, the Overall Lab is at the forefront of research aimed at improving the lives of millions of Canadians and decreasing the financial burden of dental caries and periodontitis on Canada’s health system.
Contact:
Prof. Chris Overall
Department of Oral Biological and Medical Sciences, University of British Columbia
2350 Health Sciences Mall, Room 4.401
Vancouver, BC V6T 1Z3, Canada
Tel: +1-604-822-3561
Mail: chris.overall@ubc.ca
Web: www.clip.ubc.ca
Facebook: https://www.facebook.com/overall.lab
Twitter: https://twitter.com/OverallLabNews
Further reading:
1U. Eckhard, G. Marino, S.R. Abbey, G. Tharmarajah, I. Matthew, C.M. Overall, The Human Dental Pulp Proteome and N-Terminome: Levering the Unexplored Potential of Semitryptic Peptides Enriched by TAILS to Identify Missing Proteins in the Human Proteome Project in Underexplored Tissues, J. Proteome Res. 14 (2015) 3568–3582. doi:10.1021/acs.jproteome.5b00579 or PDF download.
2Y.-K. Paik, G.S. Omenn, C.M. Overall, E.W. Deutsch, W.S. Hancock, Recent Advances in the Chromosome-Centric Human Proteome Project: Missing Proteins in the Spot Light, J. Proteome Res. 14 (2015) 3409–3414. doi:10.1021/acs.jproteome.5b00785 or PDF download.
3G. Marino, U. Eckhard, C.M. Overall, Protein Termini and Their Modifications Revealed by Positional Proteomics, ACS Chem. Biol. 10 (2015) 1754–1764. doi:10.1021/acschembio.5b00189 or PDF download.
4C. López-Otín, C.M. Overall, Protease degradomics: a new challenge for proteomics, Nat. Rev. Mol. Cell Biol. 3 (2002) 509–519. doi:10.1038/nrm858.
5A. Prudova, K. Serrano, U. Eckhard, N. Fortelny, D.V. Devine, C.M. Overall, TAILS N-terminomics of human platelets reveals pervasive metalloproteinase dependent proteolytic processing in storage, Blood. (2014) blood–2014–04–569640. doi:10.1182/blood-2014-04-569640.
6P.F. Lange, P.F. Huesgen, K. Nguyen, C.M. Overall, Annotating N termini for the human proteome project: N termini and Nα-acetylation status differentiate stable cleaved protein species from degradation remnants in the human erythrocyte proteome, J. Proteome Res. 13 (2014) 2028–2044. doi:10.1021/pr401191w or PDF download.
7U. Eckhard, G. Marino, S.R. Abbey, I. Matthew, C.M. Overall, TAILS N-terminomic and proteomic datasets of healthy human dental pulp, Data Brief. 5 (2015) 542–548. doi:10.1016/j.dib.2015.10.003 or PDF download.
8Y.-K. Paik, S.-K. Jeong, G.S. Omenn, M. Uhlen, S. Hanash, S.Y. Cho, et al., The Chromosome-Centric Human Proteome Project for cataloging proteins encoded in the genome, Nat. Biotechnol. 30 (2012) 221–223. doi:10.1038/nbt.2152.